Genetics of domestication
Motivation: Darwin's view on domestication
In the first chapter of the book On the Origin of Species (1859), Darwin mentioned the importance of so-called sports, which are mutants that show discontinuous variation, in animal and plant breeding.
For him, they are useful to explain the existence of aberrant mutations that may not be useful in nature but are selected for breeding in humans (Figure 1).
The domestication syndrome
The historical process of crop domestication has resulted in crop plants that in many aspects look very different from their wild ancestors. These differences are the result of artificial selection by humans and reflect changes in the genetic architecture of traits causing the transition from a wild plant species to a cultivated crop.
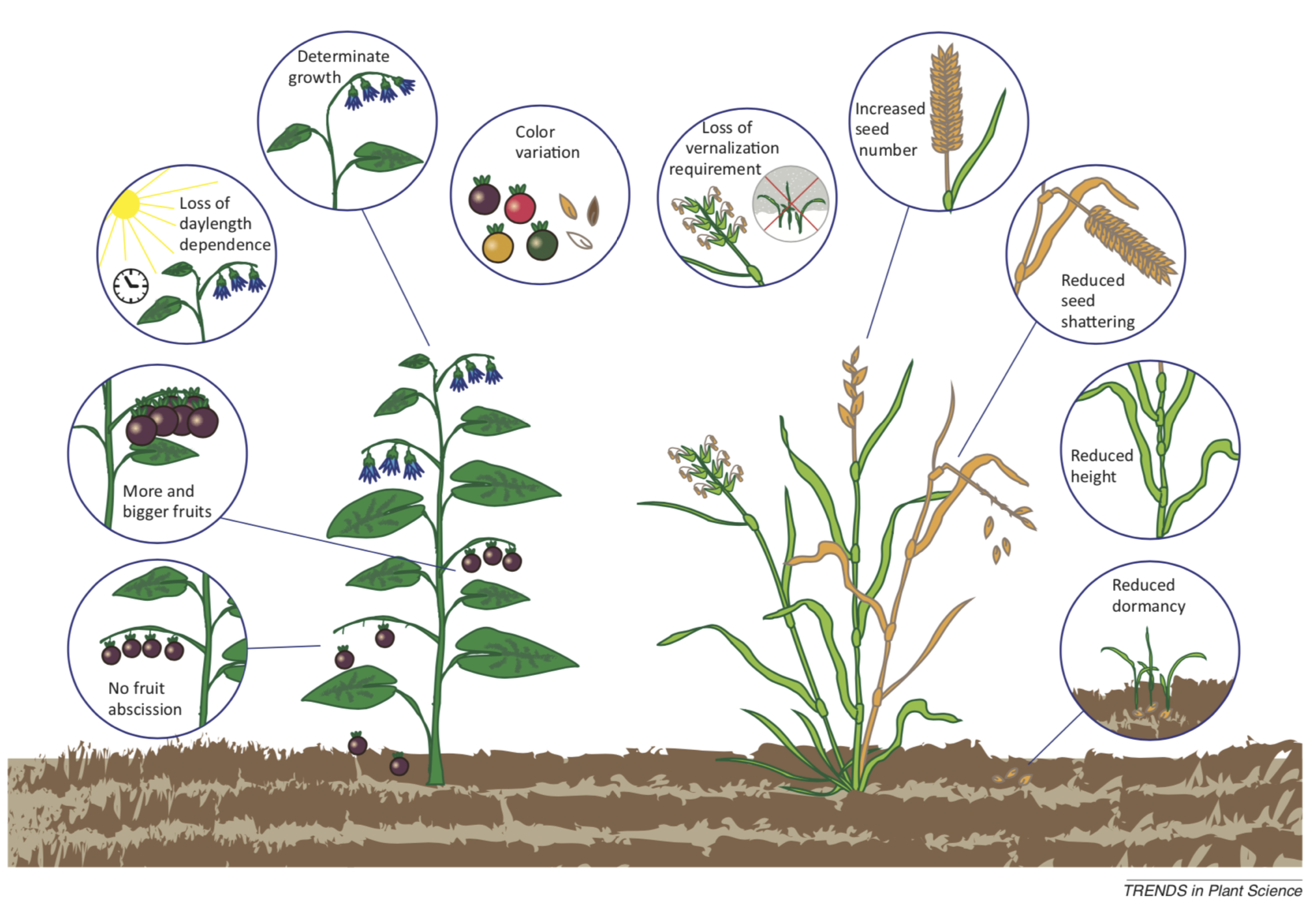
Although very different types of plants were domesticated, such as cereals, potatoes and, for example, pulse crops, similar traits and adaptations to plant cultivation were selected in these domesticates. It is noteworthy that all domesticated crops have closely related species that have not been domesticated. The combination of genetic traits shared by successfully domesticated crops is called domestication syndrome. Since the number of key domestication traits is limited, it is generally believed that only a small number of genes is responsible for the domestication syndrome of a species.
A list of key domestication traits is shown in Table Table 1. Any crop does not necessarily have all domestication traits, and they also depend on the particular farming system. For some crops such as cereals, trait size is very important, therefore all domesticates have larger seeds than their ancestors, whereas in tuber crops such as potato, reduced toxin content is more important than seed size.
| Trait | Wild Plant | Domesticated crop |
|---|---|---|
| Height | Tall | Short or dwarf |
| Growth habit | Branched and bushy | Unbranched and compact |
| Ripening | Asynchronous | Synchronous |
| Seed dormancy | Present | Absent |
| Seed shattering | Shattering heads | Nonshattering heads |
| Seed size | Small | Large |
| Ease of dispersal | Highly dispersible | Loss of dispersal |
| Threshing | Hard | Easy |
| Reproduction | Outbreeding | Self-fertilizing |
| Germination | Asynchronous | Synchronous |
| Hairs and/or spines | Present | Absent or reduced |
| Toxins | Present | Absent or reduced |
The different domestication traits can be summarized with respect to three major characteristics:
- Seeds: Reduced dormancy and increased germination ability and vigor
- Harvest: High yield, no seed dispersal and synchronicity
- Adaptation: Environment, cultivation and use
Some domestication traits differ between monocots and dicots, others are similar (e.g., larger seed and fruit size). Figure 2 shows a comparison of the most important aspects of the domestication syndromes in both taxonomic groups. For many of these traits, causal genes were already identified, and a list of such genes is provided by Lenser and Theißen (2013).
Examples of domestication syndromes are shown in the following sections.
Examples of crop domestication
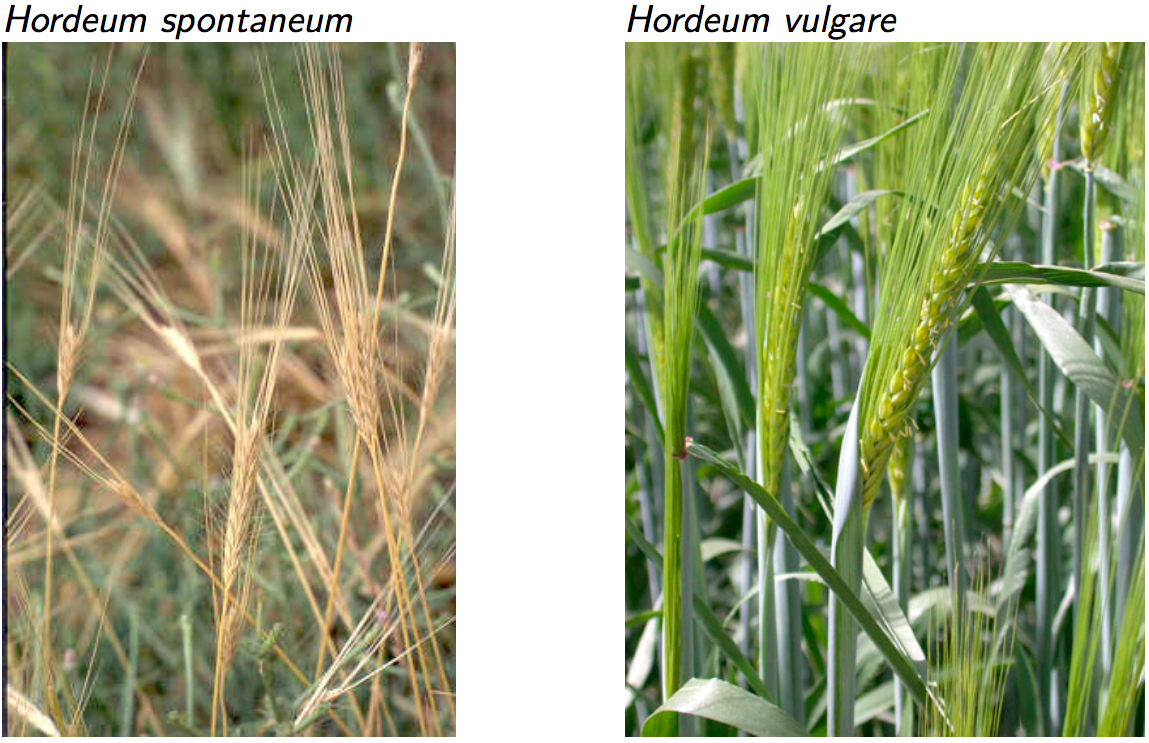
Domestication of barley
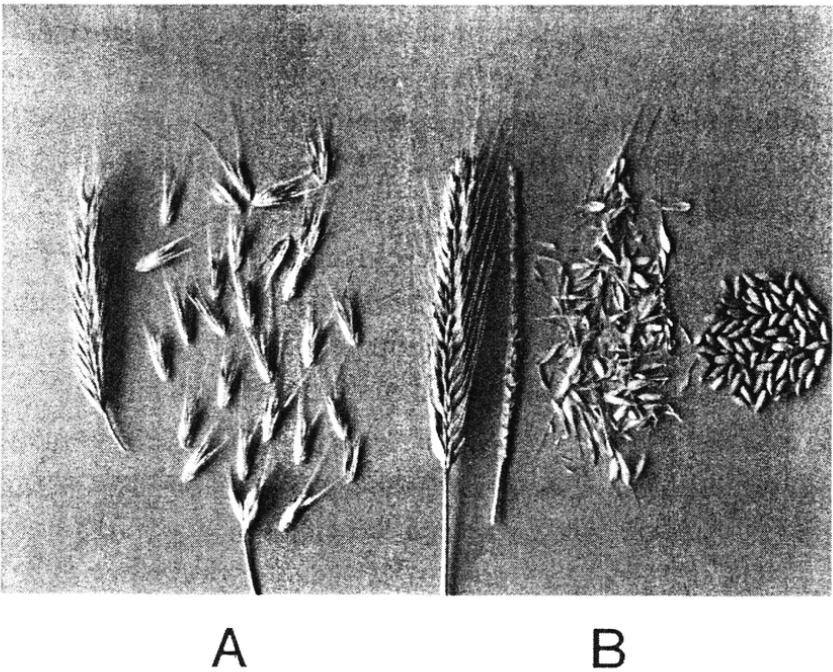
Wild barley (Hordeum spontaneum) is the wild ancestor of cultivated barley (Hordeum vulgare). The plant phenotypes are shown in Figure 3, and the awn phenotypes in Figure 4.
In wild barley, the awn shatters its seeds, and the axis of the awn falls into individual segments, whereas in cultivated barley the seeds remain attached to the awn (non-shattering phenotype), and after manual removal of the seeds, the awn axis remains intact. In contrast to wild barley, cultivated barley is also independent of the photoperiod.
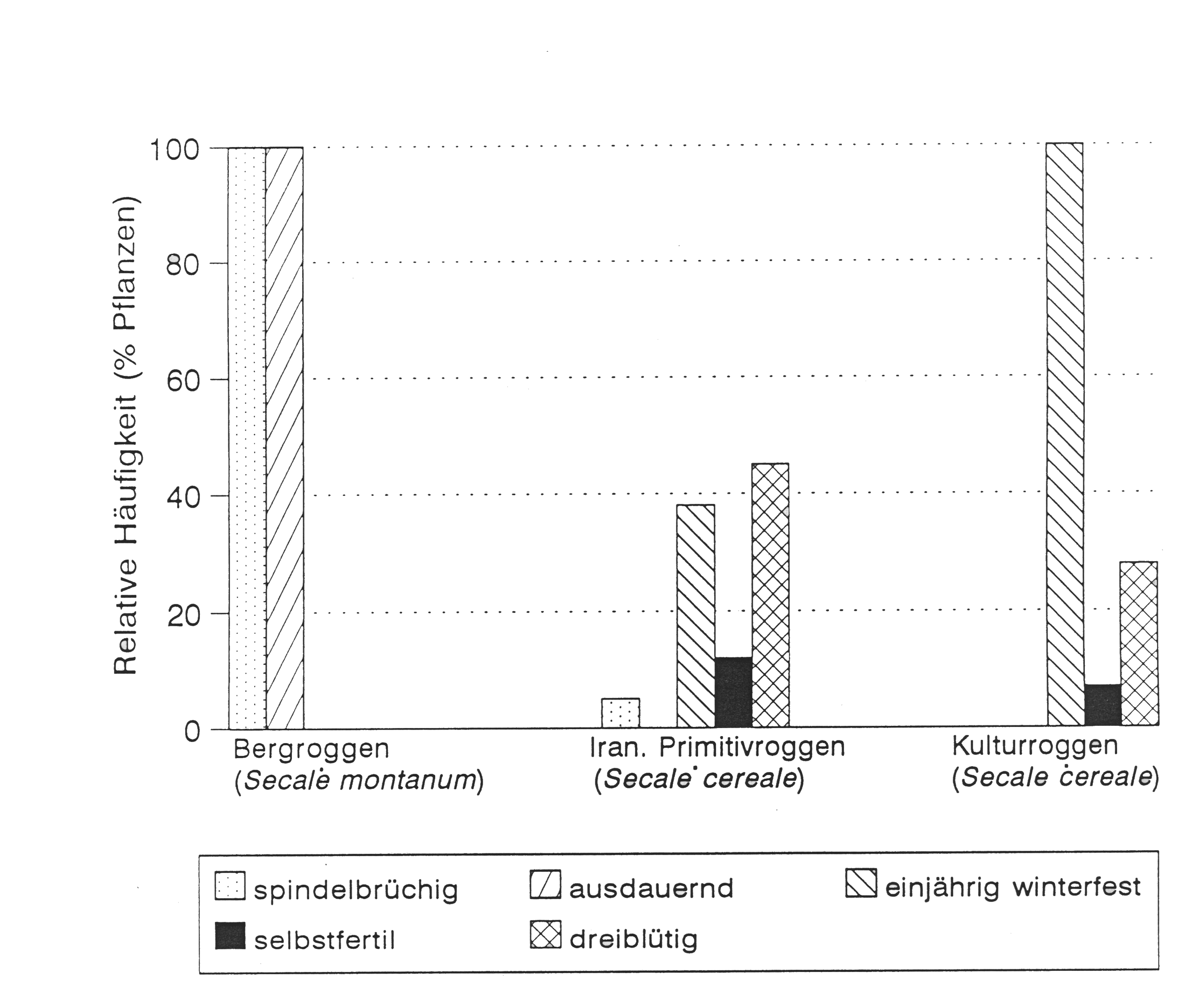
Domestication of rye
Similar to barley, the most important domestication trait of rye is seed shattering. However, because of the outcrossing nature of rye, and the high level of gene flow between wild and cultivated rye, many (domesticated) landraces are still shattering to some degree.
Only modern elite varieties are completely fixed for the non-shattering allele. Other traits that are associated with domestication are different levels of self-fertility, an annual versus perennial lifestyle, winter hardiness and flower morphology (Figure 6).
Domestication of maize from teosinte
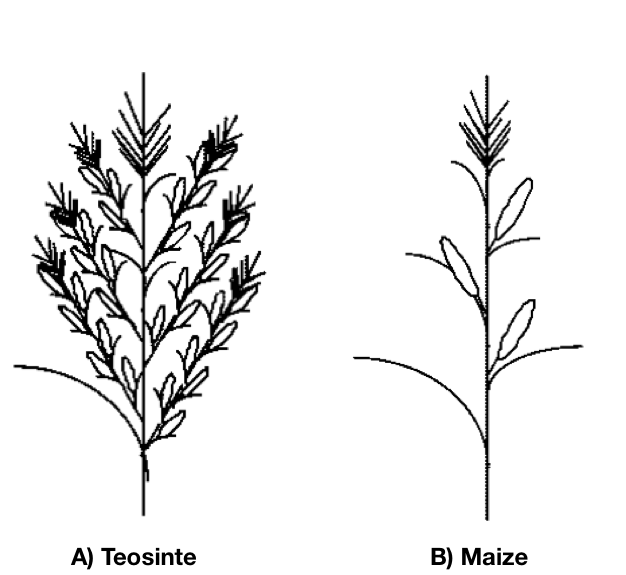
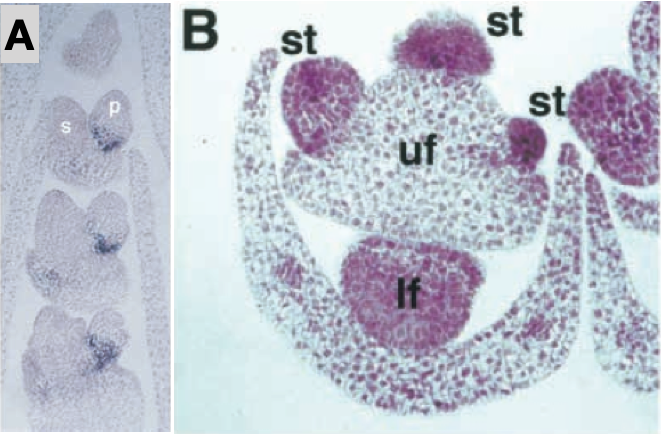
The domestication of maize from teosinte has involved some major changes in the plant architecture. These are among the most extreme differences between a wild ancestor and a domesticated crop. For this reason, taxonomists thought for a long time that maize and teosinte represent two different species. One example of the differences are the number and arrangement of male and female inflorescences (Figure 7).
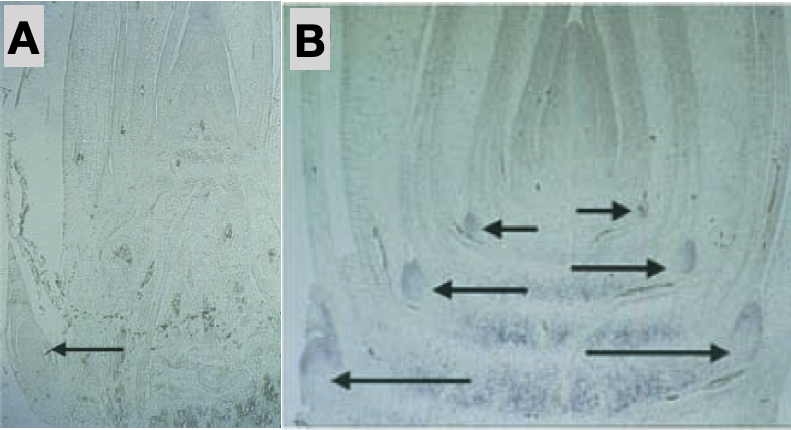
In addition, the number of shoots differs between teosinte and maize. Teosinte develops several shoots, whereas in maize only a single main shoot develops (Figure 8).
Genetic studies, however, indicated that their genomes are highly similar and that the two 'species' can be crossed to produce fertile F\(_1\) offspring.
Of the four known teosinte species, the subspecies Zea mays ssp. parviglumis is most closely related to cultivated maize and most likely the direct ancestor.
The female inflorescences or ears are very different. In teosinte, they harbor only 5-12 kernels that are covered by a hard casing. These fruit cases shatter upon maturation and are the units of dispersal. Maize ears can have up to 500 kernels, which are firmly attached to the central axis (spindle) even after maturation. They are naked and can be eaten by animals. The the ear falls to the ground, all grains mature within a short local region, shade each other and prevent maturation and flowering. Therefore, maize is entirely dependent on humans.
| Trait | Teosinte | Maize |
|---|---|---|
| Kernel number | 5-12 kernels | 500 kernels |
| Seed protection | Hard casing | Naked |
| Central axis | Absent | Present |
| Attachment to axis | - | Firmly attached |
These differences raise the question of how complex the genetics of domestication was and how many genes are responsible for the differences between teosinte and maize. There are four key traits that differentiate both:
- Each kernel is completely covered and protected by a hard, cupulate (cup-like) fruitcase consisting of a glume and an invaginated rachis segment. They are still present but strongly reduced in maize.
- Teosinte ears disarticulate and shatter upon maturation, whereas in maize mature kernels remain attached to the ear.
- Each cupule fruitcase holds a single spike (kernel-bearing structure) in teosinte, but two in maize. In teosinte, the second one is present, but is aborted during development. Hence, in maize it is de-repressed.
- In teosinte, cupulate fruitcases are arranged opposite in two rows. In maize they are arranged in four or more ranks (also called rows).
However, it was later noticed that the architecture of the two plants are also different. In teosinte, each primary lateral branch is elongated and has a tassel on its tip (Figure 7). In maize, each lateral branch is short and has an ear on its tip. In teosinte, there are secondary lateral branches terminated by a tassel, which are the male inflorescences. In maize, the lateral branches terminated by a tassel are uncommon. The leaf morphology between the two types is also different.
Identification of domestication genes
Given the large number of differences in the domestication traits the question arises how many genes contribute to the domestication syndrome.
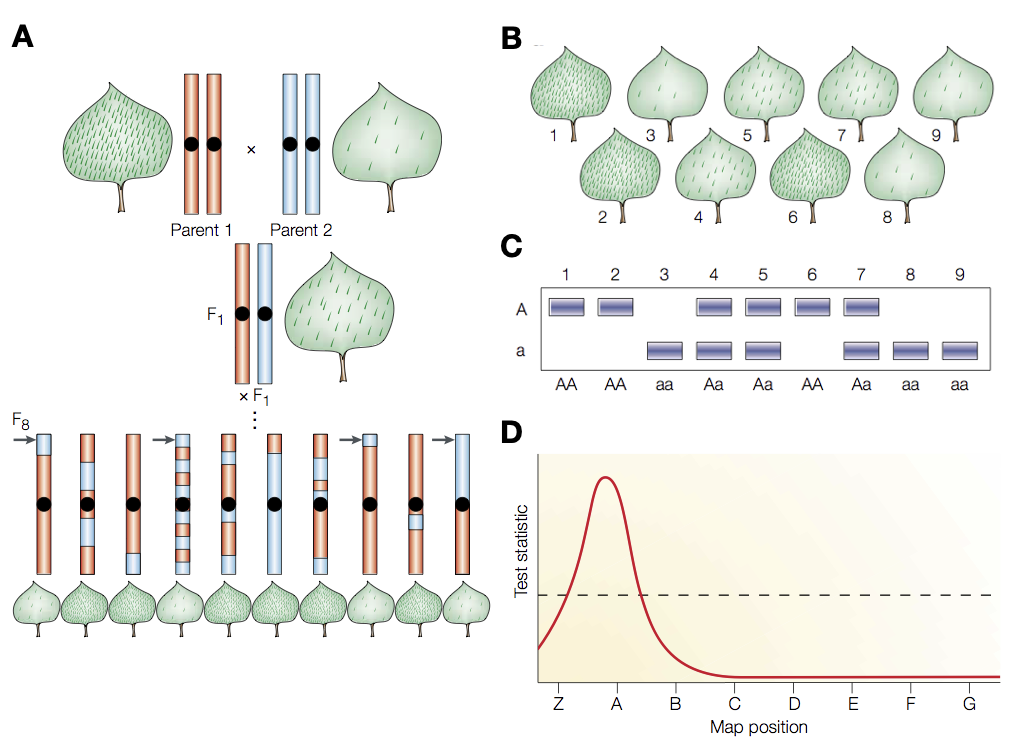
A classical approach to achieve this goal is quantitative trait loci (QTL) mapping. In this method, a cross between an wild ancestor and a domesticate are made (e.g, between teosinte and maize) and the offspring (usually the F2) generation is phenotyped for traits, which are segregating among the offspring.
Figure 10 shows an outline of QTL mapping. Two parents that differ in key traits are crossed and propagated via the hybrid F1 generation to the F2 generation. Due to recombination in the F1 generations, segments of the two parental genomes are admixed and distibuted essentially randomly onto the individuals of the F2 generation. The individuals in the F2 generation are then both phenotyped for the traits of interests, and genotyped for genome-wide genetic markers. Subsequently, a statistical test for the association between genotypes and phenotypes is conducted to identify regions in the genome which harbor genes influencing the trait.
The genetic basis of these four traits was investigated by crossing maize with teosinte to determine the number of genes controlling each trait by a QTL analysis of the offspring of these crosses. By analyzing F\(_2\) mapping populations, the group of John Doebley found QTLs scattered throughout the genome, but five to six regions had a particularly strong effect on the phenotype (Doebley and Stec, 1993). For individual key traits, up to 10 QTLs were identified that differed between the two mapping populations used. This indicated a complex genetic architecture with characteristics of quantitative rather than Mendelian inheritance.
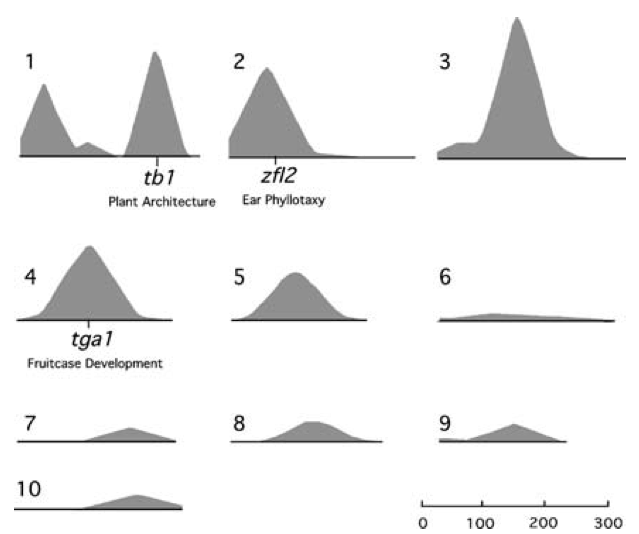
In a later review of his work , among 9 traits that influence plant and infloresence architecture, only a small number of strong QTL effects were identified (Figure 11).
The teosinte branched1 (tb1) locus in maize
One of the genes controlling a QTL for the architecture was identified as the teosinte branched1, tb1 gene (Doebley et al., 1997). Maize plants that have the teosinte allele (tb1-ref) have long terminal branches tipped by tassels, similar to teosinte (Figure 12). They also have many tillers at the basal nodes. But there are also differences to teosinte. tb1-ref plants do not form normal ears, and their secondary branches typically show only sterile, tassel-like inflorescences where teosinte bears its ears (Figure 12 a). tb1 can be viewed as an apical dominance mutant, in which homozygous mutant plants exhibit unrestrained outgrowth of the axillary meristems.
The tb1 gene was also characterized on the molecular level (Hubbard et al., 2002). It has a complex expression pattern during plant development and is a trancriptional regulator that (in its functional mode) acts as a repressor of growth. The mutation that differentiates teosinte and maize tb1 alleles are in the regulatory region and not in the protein-coding regions of the gene. The main difference between teosinte and maize is that tb1 is not expressed at the shoot apex meristems in teosinte, but in maize.
Interestingly, the expression of tb1 in other tissues such as the female spikelet is not changed between maize and teosinte.
Further analyses revealed that tb1 is a member of the TCP family of DNA-binding transcriptional regulators (Kosugi and Ohashi, 2002). It acts a repressor of transcription.
A short introduction to transcription factors: They are group of proteins that can bind to DNA in transcription factor binding sites, which are upstream regulatory regions of genes, and influence transcription of target genes. Transcription factors can activate or repress transcription and are then called either activator or repressor, respectively. Transcription factor families are classified by DNA-binding protein motifs:
- Helix-turn-helix proteins
- Zink-finger proteins
- Leucin zipper proteins
- Helix-loop-helix proteins
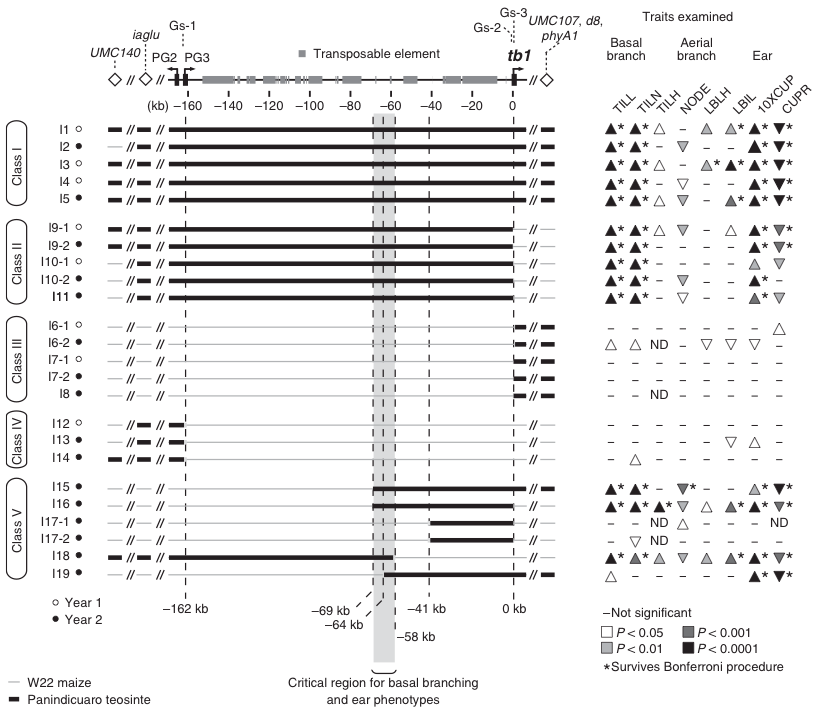
To understand the causal domestication mutation, the tb1 regulatory region was dissected molecularly. The causal mutation is located in a genomic intervall of 160 kb between two genes, pg3 and tb1. The identification of the causal mutation was achieved by introgression of a teosinte fragment into maize by backcrossing and fine mapping.
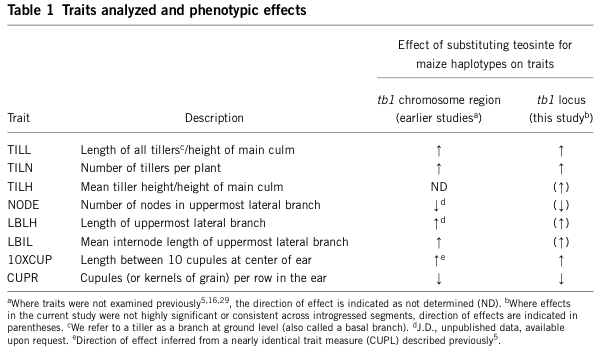
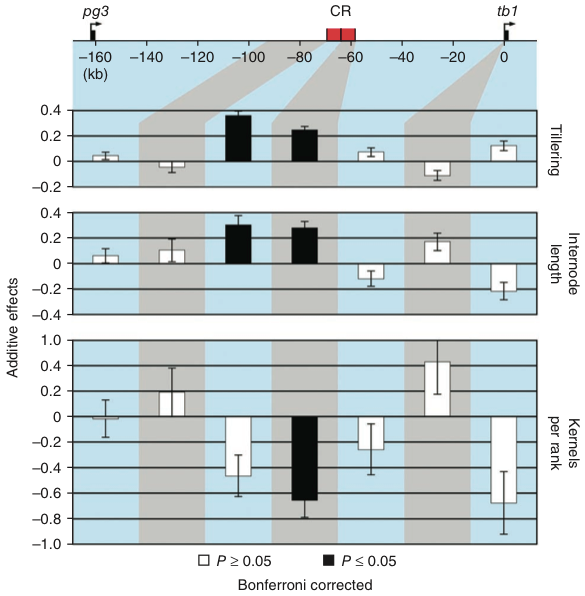
Table Table 3 shows the effects of the teosinte segment containing the teosinte allele of the tb1 gene into maize. The introgressed teosinte segment changes the tiller length and tiller numbers towards the teosinte phenotype, as expected. This work also proves that tb1 has a very specific effect on a domestication trait.
Subsequent fine-mapping defined the causal mutation to a region of 10 kb in the genome. Fine-mapping is achieved by using more markers in a genomic target region, and by examining a larger number of crosses, to investigate more recombination events.
After the fine-mapping, additional work was necessary further zoom in into the candidate gene. Without going into technical detals the approach was to make the teosinte introgressions isogenic, evaluate the resulting genetic lines in field trials and estimate additive genetic effects on the phenotype with linear mixed models.
The main results of the work was that two independent components (CR region in Figure 16) affect the tb1 phenotype and that these components resemble a distal and proximal part of the 10 kb control region, which both have positive additive effects on the phenotype.
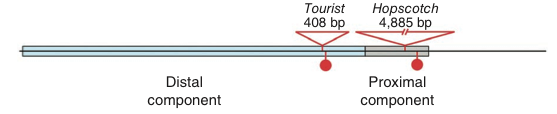
Comparison of the genetic diversity of the control region in multiple accessions of teosinte and maize genotypes identified four fixed differences, of which two are point mutations and two are transposon insertions in maize.
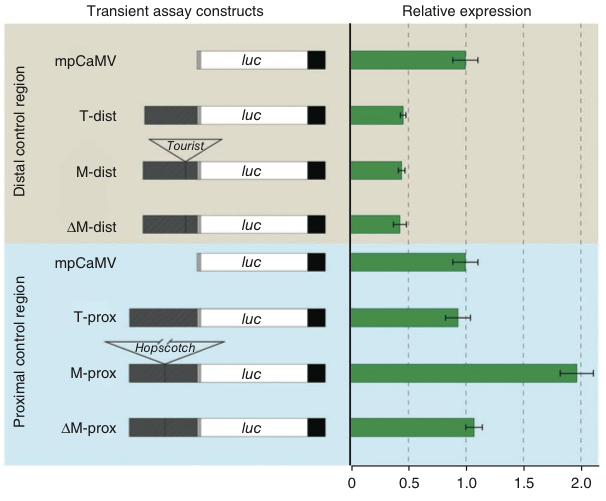
Using a so-called transient assay of the control region, gene expression of a reporter gene (luc) of different parts of the control region were determined. In this assay, a the expression of a reporter gene was driven by pieces of the functional regulatory region (CR) identified in the fine-scale mapping. The results show that gene expression of the reporter gene is increased only by the maize proximal fragment, which contains a Hopscotch transposable element (M-prox) (Figure 18).
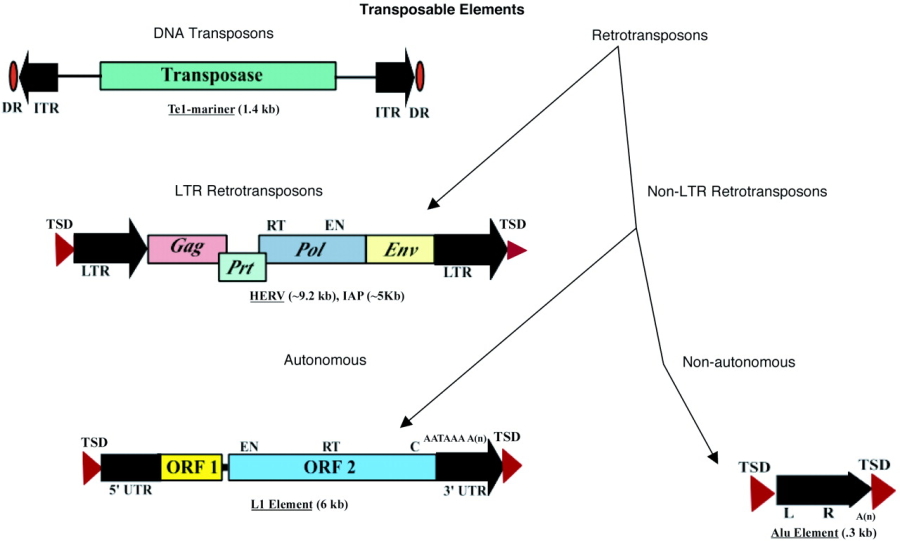
Transposable elements (TE) were discovered by Barbara McClintock (Wikipedia) in the 1940s in Maize. She received the Nobel prize for this discovery. There are multiple classes of TE, which differ in structure and the mechanism of transposition (Figure 19) TEs that require reverse transcription (transcription of RNA to DNA) are called Retrotransposons. TEs that use a transposase enzyme for insertion and excision are DNA transposons. TEs also differ with respect to their mechanism of proliferation. Autonomous TEs transpose themselves, whereas Non-autonomous TEs need other TEs in the vicinity to assist in transposition.
To conclude the fine mapping of the causal tb1 mutation. Both proximal and distal regions of the TE contribute to phenotype. Strongest effect are seen by the insertion of a transposable element, Hopscotch in maize. Therefore, the domestication phenotype is caused by a transposable element! Further analyses of teosinte genotypes revealed that the transposon insertion predates domestication by 10,000 years. This reflects selection by human standing genetic variation segregating at the tb1 locus. Standing genetic variation is already present for some time in a population and either neutral or nearly neutral before it becomes advantageous and is changed in frequency by selection. Fixation of standing genetic variation by selection is called soft selective sweep (Stephan, 2019). It is opposite to the selection of de novo mutations that provide higher fitness to the carrier immediately after their origin. Fixation of such mutations is called hard selective sweeps.
In summary, despite clear discontinous phenotypic differences between ancestral and domesticated plants, the genetic architecture is quantitative and complex teosinte-branched 1 is one of the best characterized domestication genes. Many domestication genes are transcriptional regulators. The causal 'domestication' mutation regulates the expression of tb1. The main effect of the domesticated allele results from a transposon insertion of a Hopscotch transposable element.
Meanwhile, genes have been cloned that control some of the other key traits that differentiate wild ancestors and their domesticates. They include, for example, Zea Floricaula/Leafy2 (zfl2) for inflorescence architecture (Bomblies and Doebley, 2006), and teosinte glume architecture 1 (tga1) for glume architecture (Wang et al., 2015).
To summarize, the quantitative genetic analysis of domestication QTLs has resulted in the discovery of a several domestication genes. It also revealed that the number of genes involved in domestication is limited, and that their genetic architecture can be complicated.
Key concepts
| \(\square\) Domestication syndrome | \(\square\) Transcription factor | \(\square\) Transposable elemenet |
| \(\square\) Transcriptional activator | \(\square\) Transcriptional repressor | \(\square\) QTL mapping |
Summary
- The set of traits that differentiate a domesticated crop from its wild ancestor(s) is called domestication syndrome.
- In the different cereal crops, many domestication traits are similar, such as reduced seed shattering.
- Most known cloned domestication genes are regulatory genes with an effect on the architecture of the traits.
- Whole genome resequencing reveals novel candidate genes for domestication.
- Genes that play a role in domestication or plant improvement can be identified by genetic mapping, or by selection scans.
- In maize a comparison of genetic mapping methods and selection scans shows that both methods identified several important domestication genes.
Further Reading
- Charles Darwin, On the Origin of Species, First Chapter. Download at http://www.gutenberg.org/ebooks/2009
- Doebley et al. (2006) The molecular genetics of crop domestication. Cell 127:1309 - Review of the molecular genetics of crop domestication
- Hartl and Clark, Principles of Populations Genetics, 4th edition, Chapters 7.1 - 7.4.
- Murphy, People Plants and Genes, Chapters 5 and 6.
- The papers cited in this lecture.
Review and discussion questions
- What is the domestication syndrome and how is it characterized? Are all crop species expected to have all characteristics of the domestication syndrome?
- What are the characteristics of 'sports' described by Darwin in the first chapter of the 'Origin of Species'? Why are they relevant for domestication and plant breeding? What would be the fate of these sports in naturally evolving (i.e., without human intervention) populations?
- What are the common and what are the different features of the domestication syndromes of barley, rye and maize?
- How does the tb1 of maize change the plant architecture?
- How can information about the genetic basis of domestication genes used for breeding of maize or other crops?
- Summarize the key features of the mutation-drift equilibrium.
- What is the relationship between the strength of selection and the region affected by genetic hitchhiking?
- What are the features of adaptive trait genes, domestication genes, yield genes, housekeeping genes with respect to expected levels of genetic variation? At which stages of natural evolution, domestication and selection in modern breeding programs are they expected to evolve under selection, and which type of selection is acting on them?
- Why are adaptive genes identified by natural selection mapping in wild ancestors interesting for plant breeding purposes?
Problems
- In a very short commentary, Doebley (2006) describes some findings on domestication genetics in other crops. What are his key conclusions with respect to general patterns of crop domestication?
- Which approaches for the identification of domestication genes are presented by Doebley et al. (2006). Which approach was most successful in their time?