Crop diversity & systematics
Motivation
The purpose of this chapter is to provide a broad overview of the diversity of crop plants and their systematic classification. An important goal is understand that certain taxonomic groups harbor more crops than others, and to discuss possible causes. We take two approaches: First we classify plants according to their evolutionary relationship, which is the taxonomic or systematic classification of plants. Second, we group crop plants by their different uses.
A third classification is possible with respect to the degree of domestication, which we define as the degree to which a crop plant has genetically changed from its ancestor. Such a classification, however, is quite difficult, and there is no good database available yet to look this information up for a specific crop.
The motivation for discussing this topic in the context of plant genetic resources is:
- that all crop plants have a history and an evolutionary linkage to wild plants. Knowledge of plant evolution allows to put things into context.
- to develop an understanding why some groups of plants have more crops than others.
- know the relevance of plant morphology and development for systematics, domestication, breeding and type of crop (e.g., grain vs. root crop).
Learning goals
To be added
The taxonomic system of plants
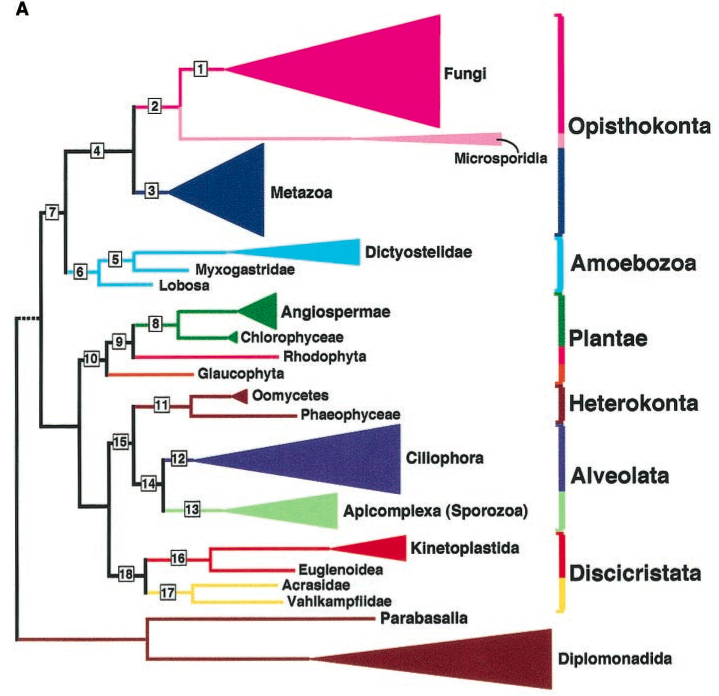
The phylogenetic analysis of the major groups of eukaryotes using a set of highly conserved genes 1 indicates that plants are grouped together with a variety of single-celled organisms (Figure 1). In contrast, fungi and animals (metazoans) are group in a different clade. A comparison of the triangles reveals that plants are not particular species-rich compared to other groups. There is much discussion regarding the age of eukaryotes, but current estimates range from 0.95 to 2.5 Billion years (Eme et al., 2014; Keeling et al., 2005). Figure 1 also shows that only a very small proportion of the total biological diversity consists of plants.
1 Highly conserved genes are present in all species in a particular taxonomic group, such as plants.
Within plants, the most important taxonomic groups are:
- Spermatophytes:
- Seed plants. They consist of the groups of gymnosperms and angiosperms.
- Gymnosperms vs. Angiosperms:
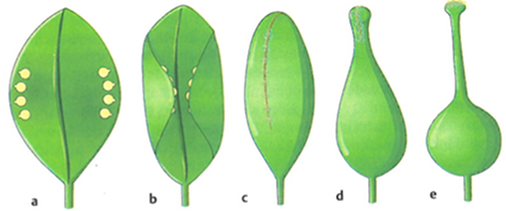
- Gymnosperms means "naked seed" and refers to the fact that their ovules (unfertilized seeds) are not enclosed and in an open condition. Gymnosperms consist mainly of the different groups of conifer trees. In contrast, the ovules and seeds of angiosperms are inclosed within an ovary (Figure 2). Another term for angiosperms is flowering plants.
- Monocotyledons vs. Dicotyledons:
- These are the two major groups of flowering plants. Monocotyledon seedlings typically have a single cotyledon (seed leaf), and the dicotyledons have two. Both groups are often abbreviated as monocots and dicots. The monocotyledons consist of 59,000 species, and they harbor the grasses (Poaceae), which are the most important family of crops. There are about 200,000 species in the dicotyledons. Essentially all field crops, vegetable species and fruit crops are either monocots or dicots.
Figure 2 shows a model for the transition of the angiosperm to gymnosperm. This model raises two questions with respect to further evolution:
- What are the expected differences in the mechanics of fertilization of gymnosperms and angiosperms?
- Why are angiosperms also called 'flowering plants'?
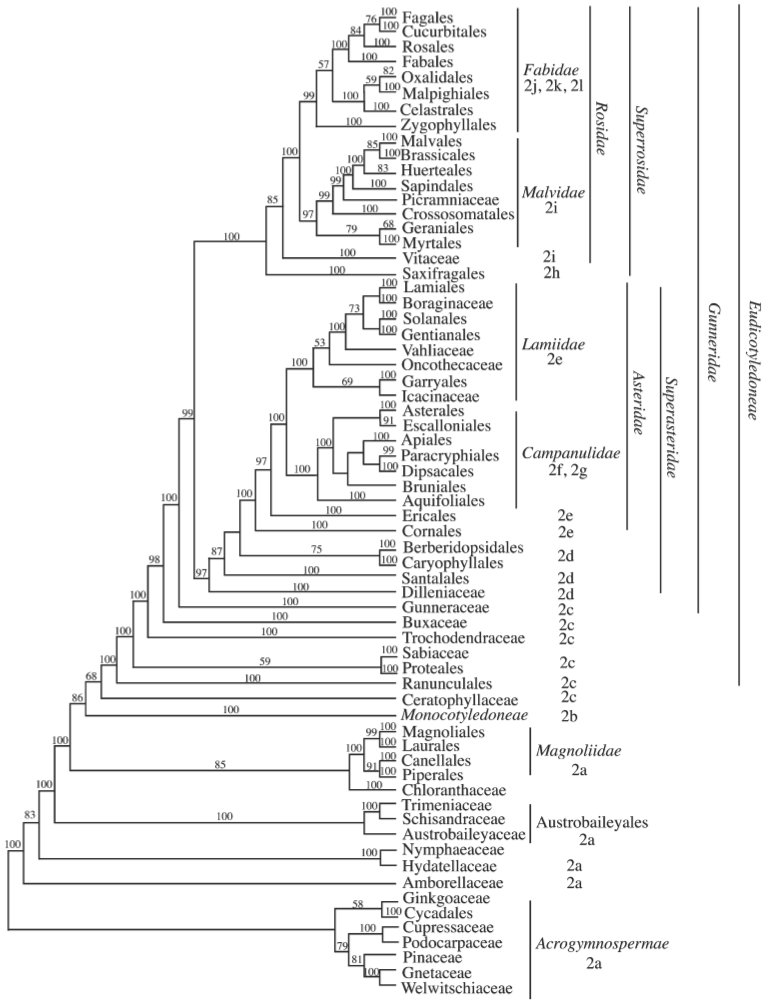
Figure 3 shows a phylogenetic tree of angiosperm plant orders and families. In this phylogeny, the evolutionary relationships of the major plant groups is shown and it can be used to compare the distribution of phenotypic traits between groups.
For crop plant, the differentiation between monocotyledons and dicotyledons is very important because both groups harbor major crops. Figure 4 shows the differences from which the name of the two groups is derived. However, there are numerous other differences between the two groups that reflect their evolutionary differences.
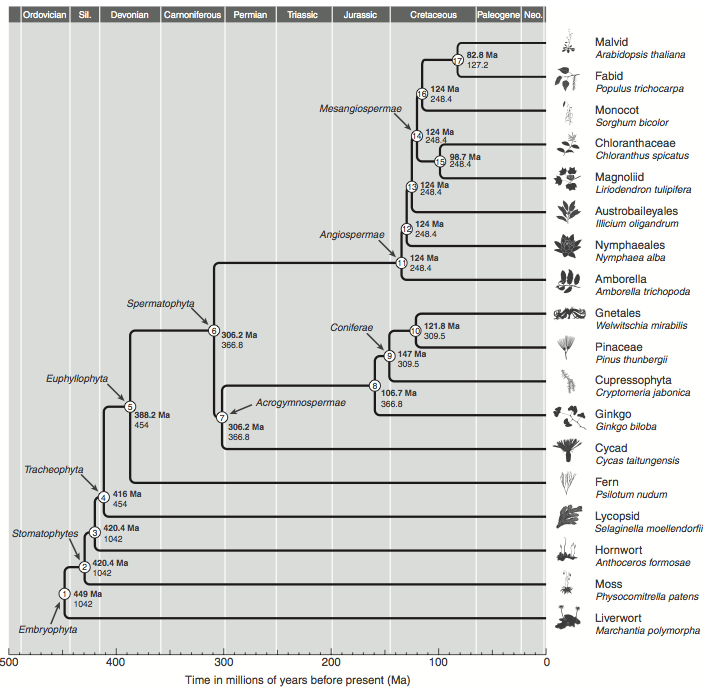
The age and the evolutionary distance between the groups is of great interest. Figure 5 shows the age of the most important groups of land plants. It shows key events in plant evolution, such as the origin of land plants, the divergence between gymnosperms and angiosperms, the divergence between monokotyledons and dicotyledons, as well as the age of various groups within the dicotyledons, such as crucifers and legumes. The common ancestor of monocots and dicots dates back 124 Mio years. However, compared to the age of the human agriculture, the events occurred in the very distant past.
Key morphological traits of crop plants
Differences in the development and morphology of traits are the basis for the phenotypic diversity of wild and crop plants. Of particular importance, some traits of crop plants have been highly modified through domestication and subsequent breeding. Most investigations of morphology and development, in particular the molecular and genetic basis of these traits and processes, were carried out with model plants such as Arabidopsis thaliana. Although many fundamental principles of plant development were discovered in this species, this model species lacks many traits that are present and important in crop species. Given the great importance of morphology and development in the breeding of crops, a thorough understanding of basic developmental processes, but also of crop-specific phenotypes, will have a great impact on future plant breeding efforts (Borrill, 2019; Schilling et al., 2018).
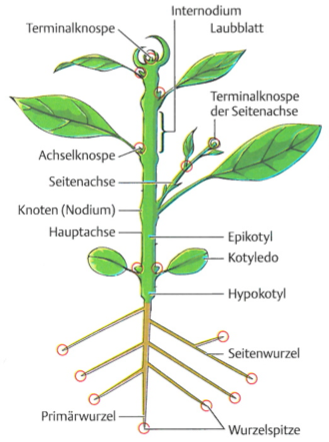
In the following, some key morphological traits are described to provide a formal context in which changes caused by plant breeding can be investigated (Figure 6).
Higher plants, as opposed to lower plants such as algae, lichens and fungi consist of three main parts:
Shoot - Leaf - Root
While this basic structure ("Bauplan") is highly conserved among higher plants, the three parts are highly variable among species, and also differ between modern crops and their wild ancestors. The structure of many plants has also been heavily modified by plant breeding.
Many review and research articles have been published that show how the targeted manipulation of developmental processes that influence (among others) morphological traits of plants can be used for plant improvement, (Boden and Østergaard, 2019; e.g., Mathan et al., 2016; Trevaskis, 2018).
Shoot
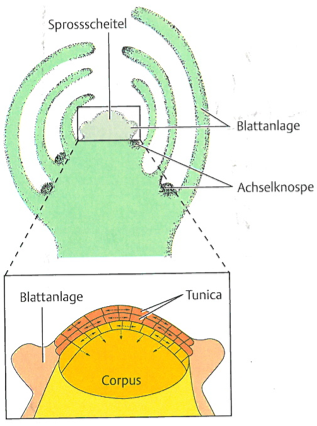
Shoots contain the shoot buds, which harbor the meristems that grow and differentiates into different specialized tissues that consist of different cell types (Figure 7). Depending on the growth patterns of shoot meristems, there are many different types of plant architecture, which frequently have been changed during plant domestication and by plant breeding.
One prominent example is the teosinte-branched 1 gene of maize, which is responsible for the conversion of multiple tillers of teosinte (the wild ancestor of maize) into a single main shoot in maize (Figure 8). The difference between the two phenotypes is caused by a mutation in the regulatory region of the tb-1 repressor gene, which causes a change in the development of apical meristems during shoot development. We will discuss the details of these phenotype in a later class on the molecular genetics of domestication.
The development of shoots is determined by genes, by the environment, and by the interaction of genes with their environment. For example, the development of a single strong shoot in some plants depends on a vernalization (i.e., cold) treatment to induce flowering, which is achieved by inducing strong apical dominance. If there is genetic variation for vernalization requirement, genotypes that require strong vernalization for apical dominance look very different from genotypes that do not require such a vernalization because the former do not achieve a state of apical dominance of the main shoot. This effect is shown in Figure 9 with the model plant Arabidopsis thaliana.
In crop plants, the propensity to form tillers (i.e., side branches) is an important trait that affects yield. The number of tillers is a key trait that differentiates maize from its wild ancestor teosinte. For this reason, the genetic and environmental control of this trait is frequently included in selection during breeding programs.
Flowers
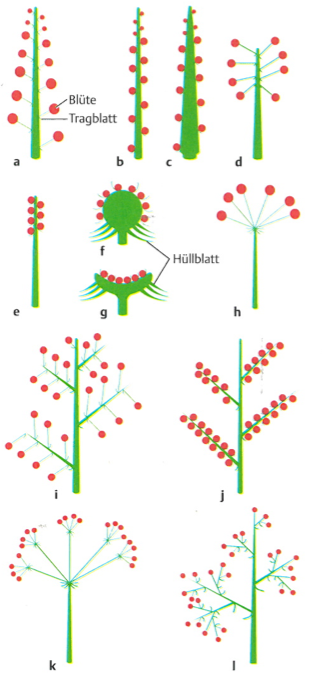
Flowering is induced by genetic or environmental factors. There are many different types of flowers. Flowers can be combined into groups, which are then called inflorescence (Figure 10)
Inflorescences can be highly diverse, and the type and arrangement of inflorescences is often changed in plant breeding and domestication. One of the most famous changes in the inflorescence is the mutation that differentiates the two-rowed from the six-rowed barley, thereby increasing the yield significantly (Figure 11).
Meanwhile, modern plant breeding efforts look for natural or artificially induced mutations that significantly change the architecture of flowers to increase yield. Artificially induced mutations can be created by treating plants with mutagenic chemicals (e.g., N-ethyl-N-nitrosourea or ENU, Wikipedia) or with irradiation (e.g., fast neutrinos), and more recently, genome editing has become the method of choice.
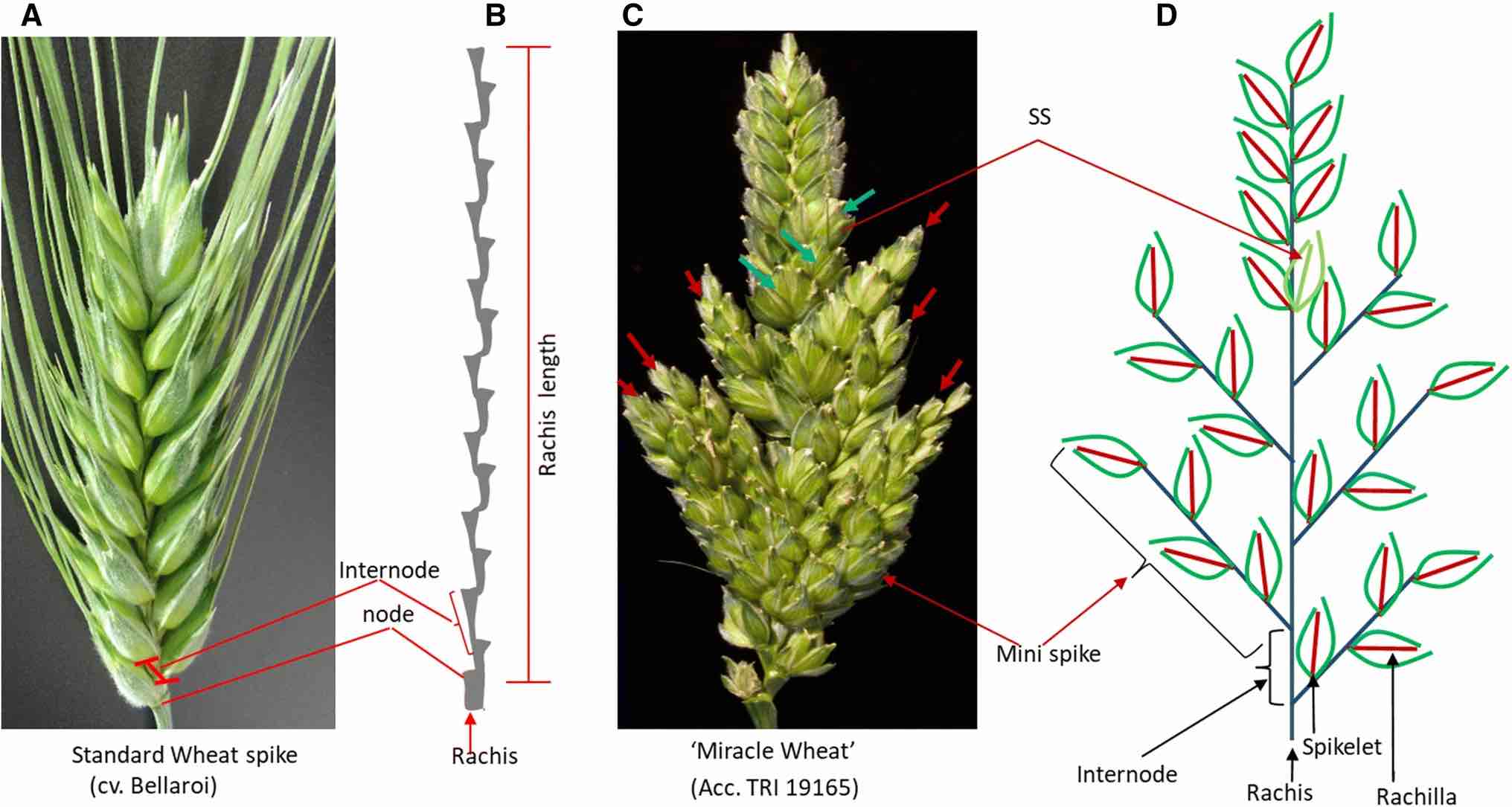
One example of a natural mutant of infloresence architecture is shown in Figure 12. The figure compares the spike of a tetraploid durum wheat elite variety with the so-called 'Miracle Wheat', which is a traditional tetraploid landrace. Genetic mapping of phenotypic variation in the offspring obtained from a cross between the two genotypes revealed that the phenotypic differences are controlled by at least nine quantitative trait loci (QTLs) (Wolde et al., 2019).
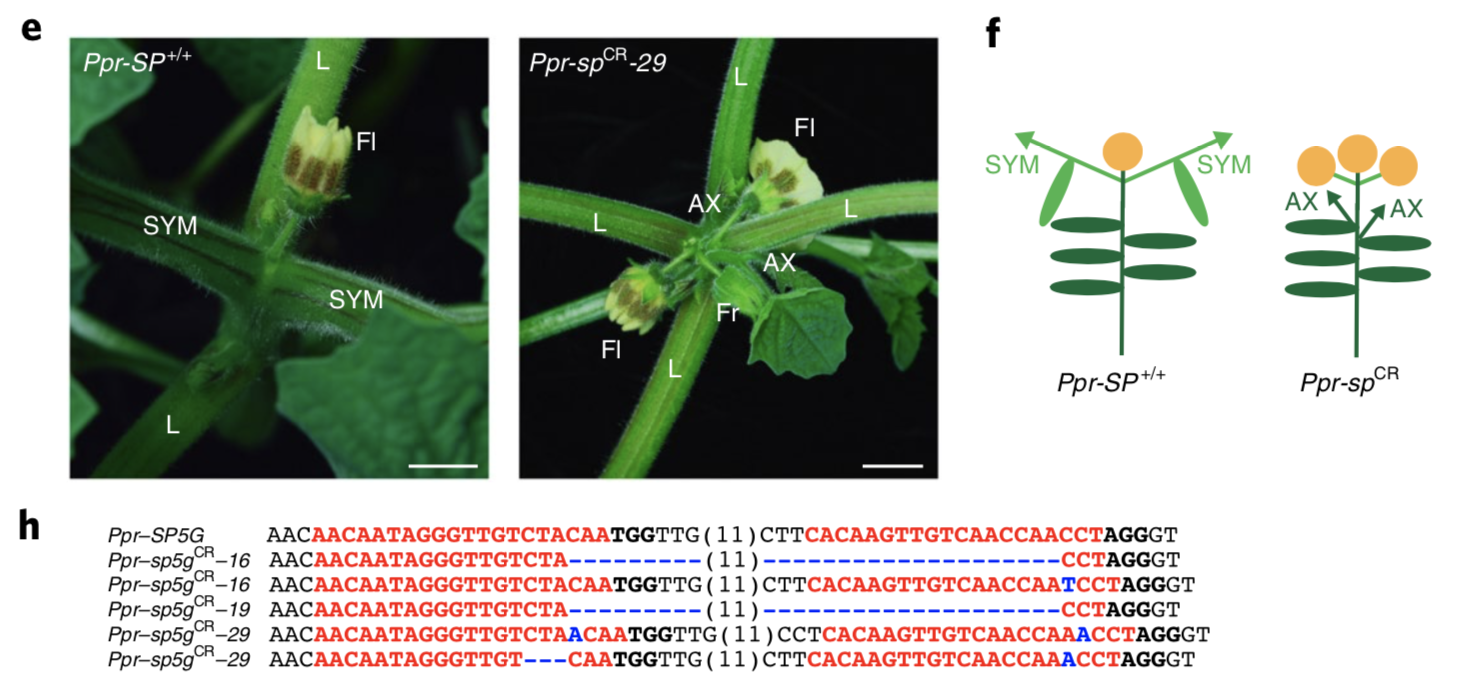
One example for the application of genome editing to change infloresence structure was shown by genome editing of the orphan (minor) crop ground cherry, Physalis pruinosa (Wikipedia) which is a close relative of tomato. A well-characterized gene, which is known to control inflorescence architecture, was mutated using CRISPR-Cas9 genome editing (Lemmon et al., 2018). Some mutations caused a change of the inflorescence architecture such that more than one fruit is produced per shoot, which increases the yield and improves the harvestability of the crop (Figure 13).
Seeds
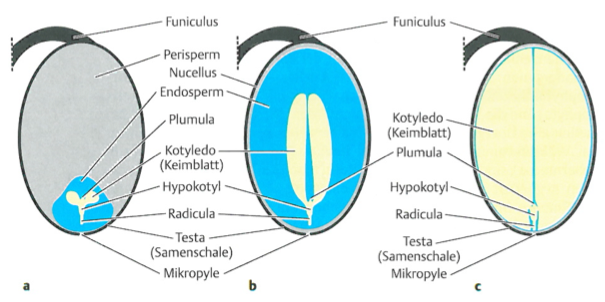
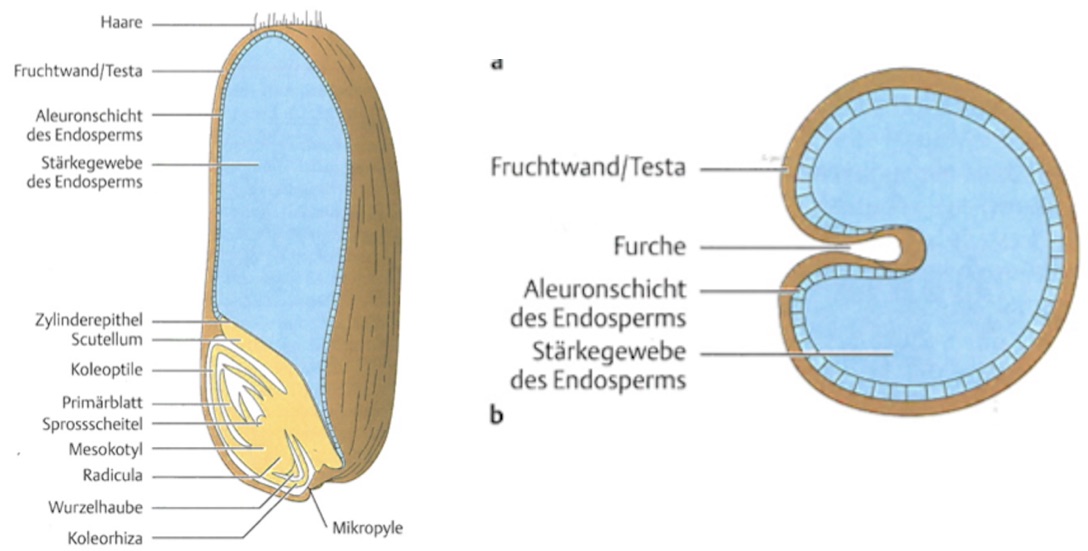
Seeds are among the most important organs of crops because they frequently are the main units of harvest. There exist different types of seeds that mainly differ by the relative proportions of perisperm, endosperm and embryo (Figure 14). Seeds are units of dispersal and long-term survival (“Dauerstage”). The seeds consists of storage tissue and the embryo. Storage material are carbohydrates, lipids, or proteins. In some species, the storage material is not in the endosperm, but in the embryo, i.e., in the cotyledons. The storage tissue supports the young seedling, until the products of photosynthesis can be utilized.
The endosperm usually surrounds the embryo, but it can also be lateral to the seeds, as for example in wheat (Figure 15)
In most crops, the perisperm is much less important than the endosperm or embryo for storage of tissues. However, in grain crops like quinoa or amaranth, the endosperm is very small and the storage tissue, which is used as food, consists mainly of the perisperm.
Roots
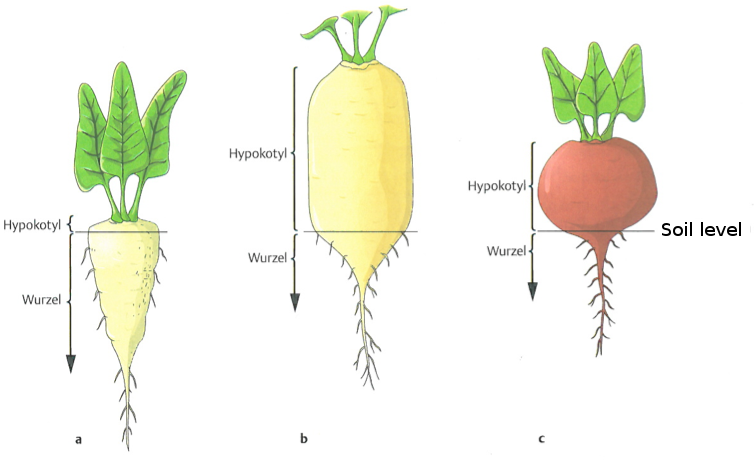
As with shoots, there are several different types of roots. Closely related crop species can differ in the type of roots as well as in the relative mass of shoot and root tissue (Figure 16).
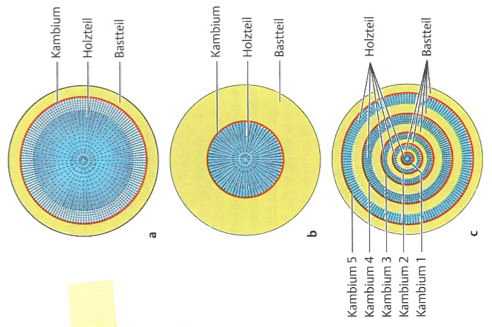
The different types of roots are generated by different numbers, polarity and relative growth rates of the meristematic growth tissues (cambium). Among closely related speces, the architecture and shape of roots can be modified substantially as a consequence of breeding and selection in order to increase the yield of the crop if the root is harvested. Figure 17 shows that not only the shape but also the composition of the different tissue types can change dramatically between closely related species.
Importance for crop evolution
The large variation of morphological traits (and associated delevopmental processes) raises the question, which traits can be modified in crop plant either by selection or genetic engineering. It is still unknown, which factors constrain variation in a given trait and which allows it. As some of the examples in this chapter have shown, this is an interesting and highly relevant area of research in crop evolution and breeding.
Distribution of crops among plant taxa
Crop plants are distributed highly unequally among the different plant families. The Mansfeld database at the German genebank is one of the best curated databases of crop plants [^2] lists about 4,800 species from 230 plant families.
The following differences between the plant families can be observed:
- Grasses and legumes harbor each more than 25% of the crop species.
- Rosaceae, Compositae, Euphorbiaceae, Labiatae and Solanaceae, all have more than 100 taxa
- Among plant families with 50 to 100 crop species there are Liliaceae, Agavaceae and Palmae.
- More than 50% of plant families harboe less than 10 crop species species.
The database contains lists of plants that are highly domesticated down to species that are hardly distinguishable from wild ancestors. The dominance of the different plant species becomes even more visible when the taxonomic distribution of the most important crops is compared (Table 1).
| Crop species | World production |
| (Mio t) | |
| Sugar cane | 1,685 |
| Maize | 844 |
| Rice | 672 |
| Wheat | 651 |
| Potatoes | 324 |
| Soybeans | 262 |
| Cassava | 230 |
| Sugar beet | 228 |
| Oil palm fruit | 211 |
| Tomatoes | 146 |
| Barley | 123 |
| Sweet potatoes | 107 |
| Bananas | 102 |
| Watermelons | 89 |
| Onions, dry | 74 |
| Apples | 70 |
| Oranges | 69 |
| Grapes | 68 |
| Seed cotton | 68 |
| Coconuts | 62 |
Why are grasses the most successful field crops?
This section discusses possible hypotheses why grasses are so important for agriculture. It is inspired by a blog post of Olivia Judson (Judson, 2010a), which was republished as a short article (Judson, 2010b). Note that the following considerations are based on hypotheses that also need to be tested.
Ecology of grasses
Much of the importance of grasses can be explained by their ecology.
- Wind-pollination
- Grasses are wind pollinated, which means that the pollen can be carried over large distances, and they are not dependend on insects for pollination.
- Fast growth
- Grasses grow fast and for this reason are rapid colonizers, for example in areas that were hit by fires or on bare soils, which is the habitat of agricultural fields.
- Resistance to herbivory
- They can withstand to being eaten because they grow their leaves from the bottom up and therefore can regenerate. Grasses frequently contain siliceous substances that are detrimental for herbivore's teeth.
- C4-Photosynthesis
- This type of photosynthesis uses less water than the C3-type of photosynthesis and is therefore more efficient in dry and hot places such as rocky outcrops or dry soils. C4 photosynthesis evolved several times independently in the grasses.
Taxonomy of grasses
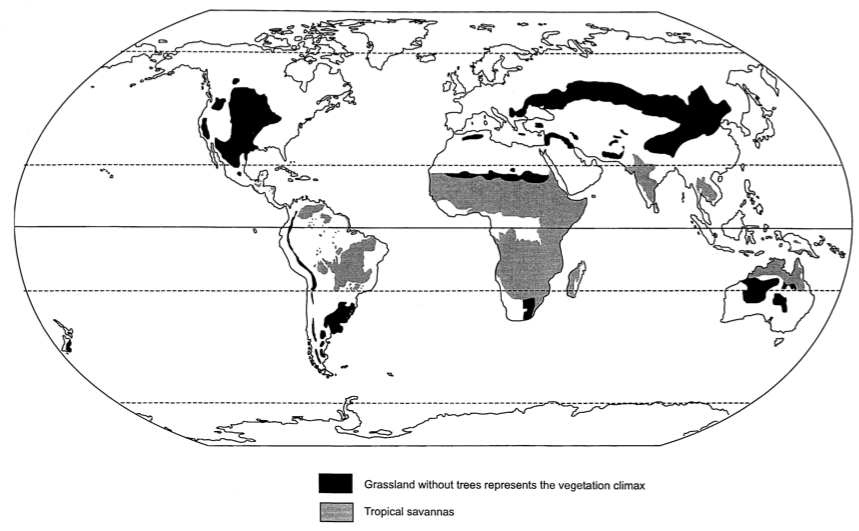
There are more than 10,000 species of grasses. More than one third of earth's biota are grasslands, where grasses are the dominant species (Figure 18). The prairie of the American mid-west is one example. Grasses show an extreme morphological diversity from small to large (bamboo).
The most important crops belonging to the grass family are: Wheat, maize, rice, barley, rye, millet, sorghum, oat and sugar cane.
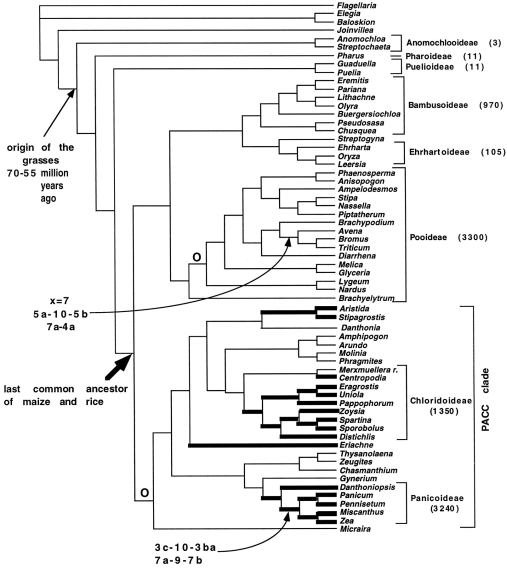
Grasses are fairly young in evolutionary terms. They originated about 80 Million years ago (Mya), shortly before the dinosaurs went extinct, and a long time after the monocot-dicot divergence, ca. 150 Mya. Grasses had a slow start, because at about 55 Mya there was little diversification, which changed until 15 Mya, when grasses were numerous and highly abundant. A phylogeny of grasses is shown in Figure 19.
Grass land create wide open spaces that led to the evolution of large herds of grass-eating animals like horses and antilopes. For this reason, grasses likely co-evolved with these animals. For example, to avoid being eaten completely, grasses produce silical opals that wear down the teeths of herbivores, which themselves adapted by developing longer and more robust or renewable teeth. Since early humans likely learned to hunt in savannahs, which is a grassland, grasses played (indirectly) an important role in the early evolution of humanity.
Key traits of grasses
Grasses have a strong and favorable effect on soils. They alter the texture of the soil, which results in smaller crumbs that harbor favorable organisms such as earthworms. The most fertile soils used now for agriculture were created by ancient grass land. An example is the prairie of North America, which was converted within a short amount of time into farmland for modern industrial agriculture.
The effect of grasses, in particular perennial grasses on the soil can be demonstrated with the intermediate wheatgrass Thinopyrum intermedium, which is about to be domesticated (Figure 20) and improved by modern genome-based breeding methods (Crain et al., 2020).
In summary, it should be noted that there is not a single explanation for the success of grasses as crops, and it seems that it is the combination of characteristics. Furthermore, numerous plant species from other groups (in particular dicotyledons) have similar characteristics but did not become major crops. For this reason, the evolutionary and agricultural success of grass species is far from fully understood.
A functional classification of crops
As an alternative to the evolutionary classification, a grouping of crop plants based on their use by humans is possible. This partitioning is taken from the book ‘Nutzpflanzenkunde’ by Lieberei and Reisdorff (2016).
- Food plants
- Plants with a technical use
The food plants are differentiated as follows:
- Use of their carbohydrates
- Use of their protein
- Use of their fat and oil
- Use of their fruits
- Vegetables and salads
- Plants for semiluxury uses (“Genussmittel”), e.g. Tobacco
- Plant-derived psyochactive drugs
- Plants with use as sweeteners
- Spices
- Fodder plants
Plants with a technical use are differentiated as follows by the substances they provide
- Fibers
- Wood
- Tannins (for tanning)
- Rubber and similar compounds
- Resin, balms and varnish
- Wax
- Dye
- Insectides
- Energy or fuel
In the current push towards a more sustainable and greener bioeconomy, plants with certain functional uses become increasingly important and also provide many opportunities for plant breeding and engineering, as well as entrepreneurial challenges.
One case in point is fiber hemp (Cannabis sativa). Although it is an old crop, only recently (through liberalisation of some regulations) (Clarke and Merlin, 2016), it has been revitalised and become a crop of great economic value because it can be used as fiber crop, for oil and food production and as medicinal and drug plant (Schluttenhofer and Yuan, 2017). This has lead to a surge of research and commercial activities surrounding this crop. As a consequence, many genomic and genetic resources, as well as experimental protocols for breeding and improvement have been generated (Kovalchuk et al., 2020; Welling et al., 2016). Such developments are frequently driven by commercial interests, and it is not common to see public-private research projects of academic researchers and companies focused on specific crops (e.g., https://newwestgenetics.com/meet-the-team) that were frequently funded by researchers.
Internet resources for crop plants
There are several databases that allow to research further information about crop plants. The most important ones are the the Mansfeld database (http://mansfeld.ipk-gatersleben.de) that contains information about essentially all crop plants of the world and contains (for each entry) links to other databases. The Global Biodiversity Information Facility (GBIF) database that contains information about wild and crop plants (http://www.gbif.org) and is currently the best maintained reference database for wild and crop plants.
Summary
- The vast majority of crop plants belong to the group of higher plants
- Higher plants are differentiated into the main groups of gymnosperms and angiosperms.
- Angiosperms are differentiated into monocotyledons and dicotyledons.
- Plants share a common basic architecture that is extensively modified among the ancestors of crops and within the crops.
- The most important groups of crops are grasses (Poaceae), which belong to the monocotyledons.
- Grasses have various characteristics that a posteriori may explain their importance as crop species.
- Crops can also be grouped by their different uses and based on their functional uses provide multiple opportunities for a bioeconomy
Key concepts
| \(\square\) Meristem | \(\square\) Shoot and root | \(\square\) Apical dominance |
| \(\square\) Cambium | \(\square\) Vernalization | \(\square\) Taxonomy vs. phylogeny |
| \(\square\) Monocot vs dicot plants |
Further Reading
- Harlan, Crops and Man, Chapters 3 and 6: General introduction into the history of crops.
- Lieberei, Reisdorff: Nutzpflanzenkunde 8th Edition, Thieme Stuttgart - Excellent overview of the diversity of crop plants and their most important traits. Unfortunately, I have not found a comparable book in english language.
- The concept of model organisms is pervasive in biological research. For comparison, you may check two short portraits of model organisms, Arabidopsis thaliana (Krämer, 2015) and Zea mays (Hake and Ross-Ibarra, 2015), of which the latter is also one of the most important crops worldwide.
- (Boden and Østergaard, 2019; Mathan et al., 2016; Trevaskis, 2018). Three different reviews on the role of plant development in crop domestication, breeding and engineering
- Soltis et al. (2019) - Concise review of angiosperms
- Kellogg (2001) - Excellent overview of grass phylogeny.
Study questions
- What is the age of the common ancestor of essentially all modern crops?
- What are likely key evolutionary advantages of angiosperms over gymnosperms? What are important consequences with respect to pollination, seed protection and seed dispersal for angiosperms?
- Which factors contribute to the fact that the number of crop plants differ between plant taxa?
- Why can it be advantageous to change the architecture (morphology and development) of crops during plant domestication and plant breeding?
- What factors determine which species of grasses were domesticated by humans?
- Why are there more dicotyledonous than among monocotyledonous, although the latter are an older group than the former?
- Which approaches could be used to determine whether a plant species can be considered a crop or not?
- What is the second most important taxonomic family of crop plants worldwide based on the number of species or production volume? Why is this the case?
- What insights can be gained by grouping crop plants according to their use?
Problems
- Legumes are the second most important family of crops. Collect some arguments from basic biology, natural and human history, breeding, agronomy, and human nutrition, why legumes have become such successful crops.
- How could the changes shown in Figures Figure 16 and Figure 17 affect agronomic properties with respect to
- Time of harvest relative to plant development?
- Mechanized versus manual harvest?
- Describe in as simple and nontechnical words as possible how the vrs1 allele in the Vrs1 gene converts a 2-rowed into a 6-rowed barley type as described in the original publication by Komatsuda et al. (2007) (See also Figure 11).
- Modern grass crops are quite unequally distributed within the grass phylogeny. What are possible explanations for this observation? Think about possible evolutionary and ecological arguments.
- Use the phylogenetic tree in Kellogg (2001) to identify the major cereal crops (and some minor crops, such as tef) and identify their evolutionary relationships. Which of the main cereals are more closely related than others? Can you find out how many million years separate the ancestors of the key cereals such as wheat, maize and rice?
A ‘personal’ crop diversity database
In order to train your data science skills, you could carry out a project throughout the whole semester by assembling and learning the key traits of the most important crops worldwide.
For this, go to the FAOSTAT database for crops (http://www.fao.org/faostat/en/#data/QC) and download the data on the 'Area harvested' for the whole world of all crops in the database ('Items' -> 'Select All') for the current year. Download the CSV file with the data, import into a spreadsheet program and sort them by the area.
For each crop add columns for the following parameters:
- Latin name
- Taxonomic family
- Place of domestication
- Main use(s)
- Lifestyle (annual/perennial)
- Diploid or polyploid? If polyploid, which type of ploidy (allo vs. auto), level of ploidy?
- Genome size; is the genome sequenced?
You can find most of the information on Wikipedia, in the Mansfield database, the GBIF database and on more specialized databases via a Google search.
In class exercise
Peer teaching
Do a roundtable and let each student in your group explain the following key terms to the other students:
| \(\square\) Meristem | \(\square\) Shoot and root | \(\square\) Apical dominance |
| \(\square\) Cambium | \(\square\) Vernalization | \(\square\) Taxonomy vs. phylogeny |
| \(\square\) Monocot vs dicot plants |
Check your knowledge about crop diversity
Briefly reflect which crop plants you ate during the last 24 hours and summarize the following information that you know from the top of your heart about these crops:
- Latin name
- Taxonomic family
- Place of domestication
- Key domestication traits
- Main use(s)
- Lifestyle (annual/perennial)
We will go through each crop in the plenum.